April 24, 2015
Evolutionary Developmental Biology, the so-called "Evo-Devo"
Embryological phenomena have different kinds of causes:
- Functional
- Genetic
- Mechanical
- Evolutionary
Highly recommended book, which was passed around in class:

None of the 4 is more fundamental than the other 3. They are interrelated. Evolutionary changes depend on genetic changes that change structures.
Historically, scientists have concentrated first on one kind of cause, then focused more on some another kind, then switched to another.
Recapitulation Ernst Haeckel
A concept based on mis-application of Darwin's evolution.
Haeckel's theories were dominant between about 1870-1900 or later.
The famous cliché "Ontogeny recapitulates phylogeny"
Meaning that the shape changes of embryonic development are speeded-up versions of changes that occurred during evolutionary history of each kind of animal.
Be careful not to open chicken eggs while they are recapitulating dinosaurs!
(I am joking.)
Recapitulation is an exaggeration; but with some truth.
Gill slits, notochord, very similar cell lineages among spirally-cleaving species: are all explainable only by shared ancestry.
If we hadn't evolved from fish, our embryos wouldn't have gill slits
Even though we now use our gill slits to form the ear canal, the tonsils, the thymus "gland", and the parathyroid gland.
(and notochords are evidence that both we and fish evolved from sea squirts).
*********************************************************
In the last couple of decades there have been big surprises about certain families of transcription factor genes that control organ formation.
For example, insect eyes have very different structures than vertebrate eyes, and their embryonic development is rather different, too!
Yet some of the transcription factors that control the locations of eye development turn out to have very similar amino acid sequences in flies as in mice, e.g. the Pax6 gene
Putting copies of mouse genes and Pax-6 proteins into flies causes them to form compound eyes in abnormal locations.
The reverse experiment also works; putting copies of fly Pax-6 genes into Xenopus frog embryos can cause them to form extra eyes at abnormal places.
Three other genes also found both in vertebrates and flies, and stimulate and are needed for eye development.
"Sine oculis", "eyes absent" and "dachshund."
But fly tissues always form compound eyes, and vertebrate tissues always form "camera eyes" (hollow, with a retina & lens)
Maybe both kinds of eyes evolved from some very simple eyes or eye-spots of flatworms? And the transcription factor proteins that stimulate expression of the rest of the eye genes stayed the same, cven though vertebrates then evolved completely different kinds of eye structures than arthropod eyes? It's anybody's guess.
What do you think?
+++++++++++++
Hearts have very different locations and structures in arthropods as compared with vertebrates (almost the reverse geometry, upside-down
Heart in the chest in vertebrates (and the neck in fish)
Neural tube runs down the back.
Arthropods (including insects and lobsters & other Crustateans)
Arthropod neural tube encircles the throat, and runs rearward down the belly
But tinman genes were found both in flies and in vertebrates
And very similar distal-less genes are expressed (locally transcribed)
- At the outer ends of fly legs
- Along the outer edge of vertebrate limb buds
- At the ends of sea urchin tube feet
- In the middle of spots on butterfly wings?
*****************************************************
Hox gene expression patterns:
Genes nearer 3' end are expressed nearer the head of the embryo.
"The colinearity phenomenon" (nobody really understands the mechanism yet)
Deletion of individual hox genes can cause one segmental structure to be replaced by an extra copy of some structure that is normally next to it.
For example: if your thumbs were replaced by an extra index finger!
(Which is NOT an actual experimental result! I just invented the possibility to dramatize the point.)
Real examples are replacement of antennae by extra legs, cnd formation of cervical vertebrae by thoracic-like vertebrae with ribs
(Cervical ribs, in your neck, can be very painful)
Hox gene effects are often "combinatorial"
Which combinations of hox gene proteins get made in embryonic tissue controls what organs develop there.
A good example is why/how snakes lost their front legs

You might have guessed that this pattern of gene expression could cause extra legs.
Question for discussion: By what mutation or experimental trick might you expect to be able to get snakes to form front legs?
+++++++++++++++++++++++++++++++++++++++++++++
The concept of "developmental constraints" (which might better have been called "evolutionary channels" or something like that)
Evolution can only produce changes that can be caused by genetic mutations (& relatively small steps at a time, all of which must produce viable body shapes.
Mechanical forces in the embryo also can only be changed little by little
THIS GAVE RISE TO AN OLD THEORY THAT VERTEBRATES EVOLVED AS UPSIDE-DOWN ARTHROPODS
What embryology has learned from fly genetics:
Revolutionary (and unexpected) discoveries came from systematic searches for genes that control early development. The biggest surprise has been that very similar sets of genes also occur in vertebrates, and almost all multi-cellular animals.The base sequences of these genes are so similar that they must have common evolutionary origins. (too similar to be convergent evolution, especially when they have different effects or functions.)
Many people conclude that the homology of genes proves that all animals must use the same basic mechanisms to create anatomical patterns as flies do, ignoring big differences such as mosaic vs. regulative development.
This is not too logical: If the genes had been discovered first in mammals,
that wouldn't be evidence that fly development is really more like mammals.
Among the bigger surprises was that the "same" genes control where eyes form
In flies as in vertebrates, even though the structures of the eyes evolved separately.
(compound eyes versus camera eyes!!)
Non-homologous organs having their locations controlled by homologous genes.
__________________________________________________________________
If mutating a certain gene causes abnormal development of a certain organ, then we conclude that the function of the protein coded by this gene must be part of the control system by which that organ is built.
A systematic search for as many different mutations as can be found that change a particular organ or function is called a genetic screen. Genetic screens have become a central part of modern embryology.
Sometimes, researchers just have to look at thousands or millions of animals to find the 2 or 3 or a dozen that are abnormal in a particular way. ("needles in a haystack")
Other times, it can be arranged for the special organisms to find themselves.
For example, if you were looking for mutations with defective phototaxis, you could plate out a few million cells, and then shine a light from one side. 99.9% of the cells would crawl toward the light; but the mutants wouldn't. The cells left behind, or moving randomly, would have a high percentage of mutants of the kind you want (plus mutants with defective locomotion).
A good screen will save you 99% + of the effort.
Some biologists' reputations (and professorships) have been earned largely by inventing a new genetic screen. Many are fiendishly clever!!
It is a huge stimulus to your cleverness to search for mutants, one by one! You get a lot of time to think of better & more powerful screens.
When you hear geneticists talking about "screens", it's probably not wire mesh.
____________________________________________________________________
With a good screen, plus a lot of effort, one can hope to identify ALL the different proteins that participate in some process, or development of some organ!
For about the last 15 years, a large percentage of researchers follow this approach.
Even when a "knock out" is an eventual part of proving the function of a gene & protein, there almost always had to be a screen to find the mutation in the first place! Elementary textbooks and courses tend to mislead about this, because it' so easy to visualize eliminating a gene (which is really done by inserting a big block of DNA into the middle of it) and then seeing the effect.
It's as if a history book said that X was discovered to be a spy because
after his execution not so many secrets reached an enemy.
That wouldn't have been the original evidence.
In brief: Geneticists developed several fiendishly clever tricks by which to "recover" recessive lethal mutations.
The first key idea is that those genes that code for proteins that control early stages of embryonic development are especially likely to be lethal when mutated.
So how do you breed dead flies?
That's where the fiendish clever genetic tricks come in.
Even harder, how do you screen for lethals that die with consistent anatomical (geometrical) abnormalities.
You want genes whose mutation produces two tails and no head,
Or that produce a head directly connected to a tail, with no middle,
Or just the anatomical structures of the middle,
Or all back and no stomach.
Or an extra pair of legs where the antennae should be,
Or two extra wings
Or extra eyes to develop from cells that should have become part of the tail.
Christine Nusslein-Volhard (a West German woman scientist) and Eric Wieschaus (a Yale Biology PhD) and several others spent ~10 years finding 140 such genes .
Incidentally, the great majority of these genes code for transcription factors.
In situ hybridization was then used to map the times & locations in developing embryos where these various genes get transcribed?
What result would you guess? If organ X fails to form as a result of mutating gene Y, then don't be surprised if messenger RNA of the normal Y gene appears in stained embryos at higher concentrations at the normal location of organ X. That's the pattern!
If your in situ stain gives that sort of result: submit a paper about it to Nature.
If you get a different result, try to make sense of it, or submit it to a lesser journal.
Mostly, results have fallen into these patterns, plus genes with very similar base sequences were then discovered in mammals, and worms, etc. even though mutating the mammal homologs produces different phenotypes Embryology is never going to be the same! It is morphing into a game of figuring out how to explain normal development is accomplished by particular combinations of normal (wild-type) alleles of these genes.
For example, the gene-that-when-mutated-causes-two-tails-and-no-heads (Bicoid) gets transcribed in the fly's mother's cell ("maternal effect"!), gets put in the front end of the fly embryos, and its translation there results in a diffusion gradient through syncytial cytoplasm that activates transcription of a series of other genes for transcription factors, different ones depending on its concentration (making it a morphogen, in Lewis Wolpert's sense of that word, and fitting theories that go back a century), and then those transcription factors bind to DNA and selectively stimulate other genes, according to which combinations of other transcription factors are present at each location (a combinatorial alternative category of morphogen!) [which some other people had hypothesized]
One family of transcription factors controls where and when transcription will be stimulated in the next (of five) families of transcription factors will be activated, much like dominos knocking over dominos, and somewhat like a cuckoo clock, designed by a cuckoo clockmaker.
But it's all true.
But maybe some aspects of development are invisible to this approach?
===========================================================
SOME MAJOR DIFFERENCES IN INSECT EMBRYOLOGY (among which fly development is somewhat special)
1) All 3 axes (anterior-posterior, dorso-ventral, medio-lateral) are already irreversibly determined before fertilization by signals and mRNA from surrounding maternal tissue.
Notice the contrast to vertebrates, especially mammals! Not to be a spoil-sport, but what are we supposed to think when the Wnt gene or some such turns out to be part of how frog eggs react to sperm entry or gravity in deciding which end should form the head?
One idea is to think of Wnt, etc. as analogous to transistors and capacitors etc. that you can find in TVs and air-conditioners and computers etc. which might lead one to conclude that these all work by the same mechanism. At some level, that would be true; at other levels false.
2) Arthropod embryos don't cleave into separate cells until the 13th mitotic cycle (~6-thousand cell stage). Until then they are syncytial, so that even fairly large proteins like transcription factors are free to diffuse around.
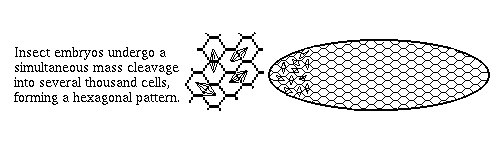
This is very different from vertebrates, and the great majority of phyla, in which nuclei are put in separate cells soon after fertilization, and before many decisions are made. Even small proteins can't diffuse from cell to cell.
3) Metamorphosis: Fly larvae shed their skins twice ("molts"),
and then during the third molt they pupate like caterpillars,
and the outer surface of the fly body is made of outpocketings
that had previously been undifferentiated sacs folded into the grub body wall.
"Imaginal Discs" Metamorphosis does not occur in many insects.
4) Most of the oocyte cytoplasm is produced in 15 mitotic sister cells, called nurse cells. These are not at all the same as follicle cells, although follicle cells perform a nursing function, and insects have follicle cells in addition to nurse cells. Vertebrates have follicle cells, but not nurse cells.
5) Mesoderm is internalized along the bottom, in an invagination process that looks like neurulation in vertebrates.
Endoderm folds inward from both ends toward the middle, In a process separate from the internalization of mesoderm.
Neural ectoderm is internalized by many separate cells, ingression over broad areas of the lower surface.
6) The ventral side is the center of embryological movements and differentiation, instead of the dorsal side as in vertebrates.
There is a very old theory, now supported by some new molecular evidence That arthropods evolved as upside-down equivalents of vertebrates, Or that vertebrates evolved upside-down, relative to arthropods.
Heart on the top; Nerve cord on the bottom!
Pipes carrying air from holes in the side, directly to each tissue!
-----------------------
What is meant by "gap genes" and the "co-linearity phenomenon".
If the normal phenotype were the alphabet as shown below:
ABCDEFGHIJKLMNOPQRSTUVWXYZ
Then the phenotype of mutants of the Bicaudal and Bicoid genes would be:
ZYXWUTSRQPONNOPQRSTUVWXYZ
Which had also been seen to develop when strings were tied tightly around the middles of dragon fly early embryos. (Two mirror-image rear halves, with no anterior organs)
________________________________________________________________
Some further complications:
First that the Drosophila hox genes are in two separate bunches on the third chromosome Which can be diagramed like this:
genegene: H1 H2 H3 H4 H5-genegenegene-H6 H7 H8 H9 H11 H12 H13-genegenegene
Second, that mammals have four different sets of 9-11 hox genes (per set) with each of the four sets each being on a different chromosome, sort of like this:
A1, A2, A3, A4, A5, A6, A7, A8, A9, A10, A11, A12, A13 B1, B2, B3, B4, B5, B6, B7, B8, B9, B10, B11, B12, B13 C1, C2, C3, C4, C5, C6, C7, C8, C9, C10, C11, C12, C13 D1, D2, D3, D4, D5, D6, D7, D8, D9, D10, D11, D12, D13
Except that 14 of them don’t exist; so the real pattern is the following in mammals
A1, A2, A3, A4, A5, A6, A7, * , A9, A10, A11, * , A13 B1, B2, B3, B4, B5, B6, B7, B8, B9, * , * , * , *
* , * , * , C4, C5, C6, * , C8, C9, C10, C11, C12, C13 D1, * , D3, D4, * , * , * , D8, D9, D10, D11, D12, D13
two lacking four lacking four lacking four lacking
The numbering is based on similarity of base sequences, so that saying that D2, D5, D6 and D7 “don’t exist” means that in the D bunch of hox genes, none are as closely homologous to A2 or B2 at those two are to each other, and similarly none in the D group have sequences as similar to A5, B5 and C5 as those three genes are to each other, etc.
What has still not been discovered...
A) What mechanism turns on transcription of these genes, at the proper locations and times in developing embryos?
B) Whether, or how, the above mechanism for spatial control of transcription uses or depends on the relative locations of the hox genes on their chromosomes?
C) How hox gene expression controls the locations of organs and tissues in the body.
Sometimes the anterior boundaries of expression of specific hox genes corresponds to particular anatomical boundaries. Examples include boundaries between fingers and toes, which correspond to anterior boundaries of expression of hox genes D13 (between little finger and ring finger, and also between the little toe and the toe next to it), hox gene D12 (between ring finger and middle finger, and also between the toes equivalent to them.), and so on for the anterior-most places where D11 and D10 are expressed, which match the locations between middle finger and index finger, and between index finger and thumb (and equivalent toes)!
In situ staining with labeled single stranded nucleic acids have proven the locations of these and other boundaries of expression of specific hox genes. What is lacking are mechanisms to find out (A above) what mechanism causes hox gene D13 to be transcribed in the tissue that becomes the little finger and little toe, but not to be transcribed in the tissues anterior to that, whereas hox genes D12 is transcribed in the little finger and in the ring finger, and D10 is transcribed in the middle finger, ring finger and also the little finger.
Nor has anybody been able to figure out (and prove) how expression of hox 13 causes the formation of the little finger. (question B above)
The best researcher on such subjects is Prof. Ann Burke, who was a member of this department for several years, until she moved to another university. One of her methods used specially engineered viruses into which certain hox genes had been inserted. By opening chicken eggs, and putting these viruses at chosen locations, she was able to change what organs would develop at those locations. Imagine causing a mammal embryo to form an extra little toe at any chosen location! She did experiments logically equivalent to that. Prof. Burke's PhD student, Julie Nowicki was a TA in this course for two years, and made major discoveries about control of anatomical patterns by hox gene transcription. She also collaborated with Prof. Alan Feduccia to test whether the 4 toes of bird feet correspond to toes 1, 2, 3, & 4, as opposed to toes 2, 3, 4 & 5, based on which D hox genes are transcribed in embryonic ostrich (!) feet. This was (very) interesting as a test of hypotheses about which particular group of reptiles birds evolved from (whether from dinosaurs, or not). Their evidence proves "not", but opposition continues.